Our Research
The physicochemical interactions between macromolecular components create a cellular machinery of great complexity. Whereas the core molecular repertoire has been identified and biochemically characterized, we understand very little about how the cell functions as a whole. To visualize and understand the orchestration of macromolecular machines we use an arsenal of biophysical technologies such as advanced microscopy techniques, high-resolution structural methods, molecular modeling, but primarily cryo-electron tomography combined with machine learning techniques. These technologies we use to understand adhesion mechanisms in both eukaryotes and bacteria. To enable adhesion the cell develop intricate structures with complex architectures. Dysfunction of those have adverse effect in maintaining cellular homeostasis and contribute to development of disease like cancer and drug resistance in bacteria.
iMOL: Interfacing Molecular and Computer Science
The ‘Interfacing image analysis and molecular life science’ (iMOL) Research Training Group (RTG) is committed to educating PhD students in the theory and practice of advanced microscopy methods and cutting-edge image analysis. Microscopy is instrumental to our understanding of the basic principles of life at multiple spatio-temporal scales and resolutions. Machine learning techniques are used to mine the information hidden in the images and recover the signal. Recent technological developments in light and electron microscopy have broadened and deepened our understanding of biological processes. This underlines the importance of and the need for dedicated educational training programs on image analysis methods that support the understanding and interpretation of microscope-derived data.
Central to Imol RTG are hypothesis-driven life science applications that challenge textbook knowledge at the molecular scale. We will emphasize on neuroscience and analyze cell-cell junctions between epithelial cells that affect the blood-brain barrier. We will also analyze how vascular and neuronal structures intermingle along the developmental process in order to generate functional organs. In the interspace of those we will also look at neurodegeneration and the effect of the formation of extracellular amyloid plaques.
In the iMOL RTG we will optimize the design, construction and automated usage of cutting-edge microscopy techniques using interdisciplinary efforts. By working with cryo-electron tomography (cryo-ET), super-resolution and light sheet-based fluorescence microscopy (e.g. SIM, PALM/dSTORM, SPIM/DSLM), Raman microscopy and multi-photon microscopy, our efforts will span the complete range of scales and resolutions, i.e. from tissues to cells and ultimately to individual macromolecules. To interpret the resulting images, we will investigate and further develop state- of-the-art computational methods and implement them on dedicated processing units (e.g. GPUs) and supercomputers for semi-autonomous image analyses and interpretations.
Dynamik von Membranen. Molekulare Grundlagen und Theoretische Beschreibung (LOEWE Dynamem)
The cells as the basic building blocks of all living organisms are, like many of their intracellular components, bounded by membranes. The membranes are therefore of fundamental importance as a spatial boundary and as a stage for biological and chemical processes and enable spatial and temporal control of cellular processes. Their biological and biophysical properties are largely determined by different lipids, lipophilic substances, integral membrane proteins and membrane-associated complexes. While the static properties of biological membranes are relatively well understood, dynamic membrane changes and the resulting physiological effects still raise major questions today.
Dramatic changes in cellular membranes are observed during the aging of biological systems, during programmed cell death, under different stress conditions, during adaptation to changed environmental conditions and during cell differentiation. A number of diseases are characterized by changes in membrane dynamics. Well-known examples are a reduced availability of synaptic vesicles or the accumulation of altered membrane structures. The influence of small molecules on membrane behavior should not be underestimated either and offers a broad field for the development of pharmaceuticals.
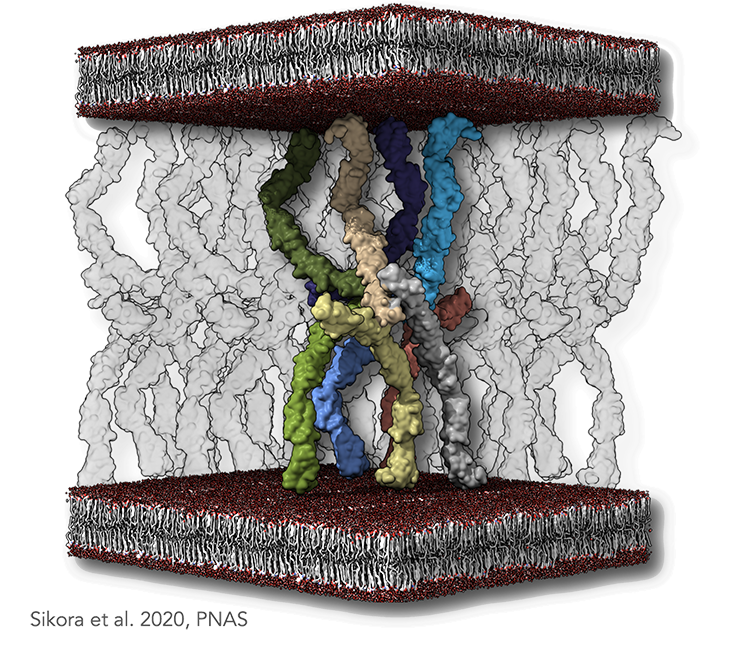
Figure legend: Desmosomal molecular architecture. (A) A minimal set of cadherins able to produce an antiparallel arrangement, with transparent molecules denoting the neighboring molecules. The plasma membranes are indicated as lipid bilayer colored in red and gray. In the dense midline plane, each node of such a lattice is composed of two types of interspersed EC1–EC2 interfaces. The first type is a cis-trans-cis interaction, as previously suggested by the X-ray structure of classical cadherins. The second type is a trans-trans-trans interaction. Both interactions are necessary to sustain the antiparallel arrangement of the cadherins.
Resistance to antibiotics, focusing on critical resistant bacteria from the WHO priority-1 list of pathogens, and resistance to antifungals
Gram-negative ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa and Enterobacter species) bacteria are increasingly successful in surviving within the hospital setting and thus a major challenge to infection treatment. They have intrinsic resistance mechanisms allowing them to survive in adverse situations, in addition to the well-documented acquired resistance to many antibiotics and chemotherapeutic agents. For the most part, the mechanisms underlying their persistence and survival success are unknown. Generally, the double-layer surface of Gram-negative bacteria, in which membrane proteins and membrane protein complexes span an inner membrane and an outer membrane (Fig. 1), appears to be central to their intrinsic resistance phenotype. The surface does not only provide a first line of defense against environmental stresses for individual cells, but enables the orchestration of higher-level defense mechanisms for multiple cells, such as the formation of biofilms. We aim to obtain an overall molecular understanding of the multiple interactive mechanisms underlying resilience in ESKAPE bacteria, with a focus on membrane protein complexes and interactions. By applying new technologies to investigate the dynamics of systems in cellulo, we expect to go even further – paving the way for the treatment of Gram-negative bacterial infections and overcoming resistance. Below, we provide a short overview of the planned research in this multifaceted project.
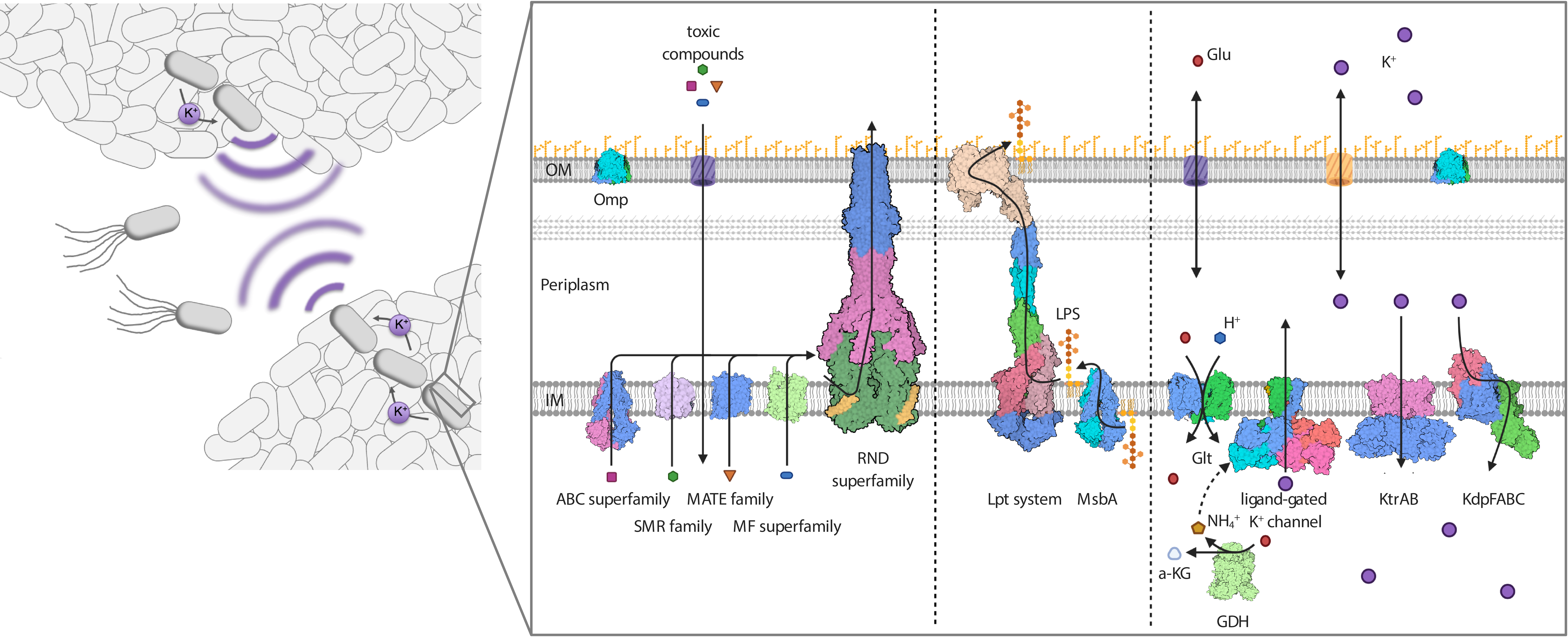
Figure legend: Protein complexes at the cell surface of Gram-negative bacteria mediate intrinsic resistance. As motile cells, but particularly when organized into biofilms, Gram-negative bacteria are resistant against antibiotics, salt stress, drought and chemical compounds such as disinfectants. Proteins of various transporter superfamilies reside in the IM and flip the antibiotics from the cytoplasm to the periplasm. From there, RND efflux pumps pump the toxic compounds over the OM. The integrity of the OM and its impermeability towards toxins, in combination with the action of the efflux pumps, result in a synergistic and powerful intrinsic resistance phenotype. The integrity of the OM critically depends on the IM transporter MsbA and the IM- and OM-spanning Lpt system (LptABCDEFG), which mediate the movement of the LPS towards the outer leaflet of the OM. Both transport systems are essential, and their inhibition is lethal to the Gram-negative cell. Resistance to salt stress and dryness, as well as biofilm formation, which is mediated by electrical signaling, rely on potassium homeostasis and the orchestrated action of potassium transporters and channels at the IM. Deletion of the individual systems significantly reduces the pathogenicity of pathogenic bacteria.
The molecular sociology of Mycoplasmas with cryo-electron tomography
Mycoplasmas are among the smallest and simplest prokaryotes capable of self-replication, with a genome which may be as small as 600 kbases. They are significant pathogens that may induce clinically relevant manifestations. We use cryo-electron microscopy to solve the structure of the main adhesion complex of the Mycoplasmas, the Nap, that is responsible for the adhesion of the pathogen to the host cell and indispensable for manifesting the infection. In addition, we studied numerous mutants of Mycoplasma genitalium by cryo-electron tomography, which allowed for explaining the mechanism of adhesion and motility. This project is developing along two avenues: (i) Using single-particle cryo-electron microscopy to solve the structure of the intracellular part of the Nap (Napic), for which we have evidence that it is responsible for triggering the adhesion release mechanism. In addition, we would like to solve the structure of other immunodominant protein on the surface of the Mycoplasmas and understand the underlying interactions with the Nap to facilitate infection. (ii) Using cryo-electron tomography and a focussed-ion-beam approach, we would like to investigate the direct interaction of the Mycoplasmas with host cells (primarily epithelial cells) in order to understand how the infection manifests. In particular, we would also like to visualize the adhesion of Mycoplasmas to epithelial cilia, which apparently causes their dysfunction during the infection. For both of the above avenues we have already done significant prior work, both independently and as members of a vibrant collaborative community. Finally, the motivation for our work is reinforced by recent studies that have reported a critical role of Mycoplasma pneumoniae for the clinical outcome of viral infections of the respiratory tract.
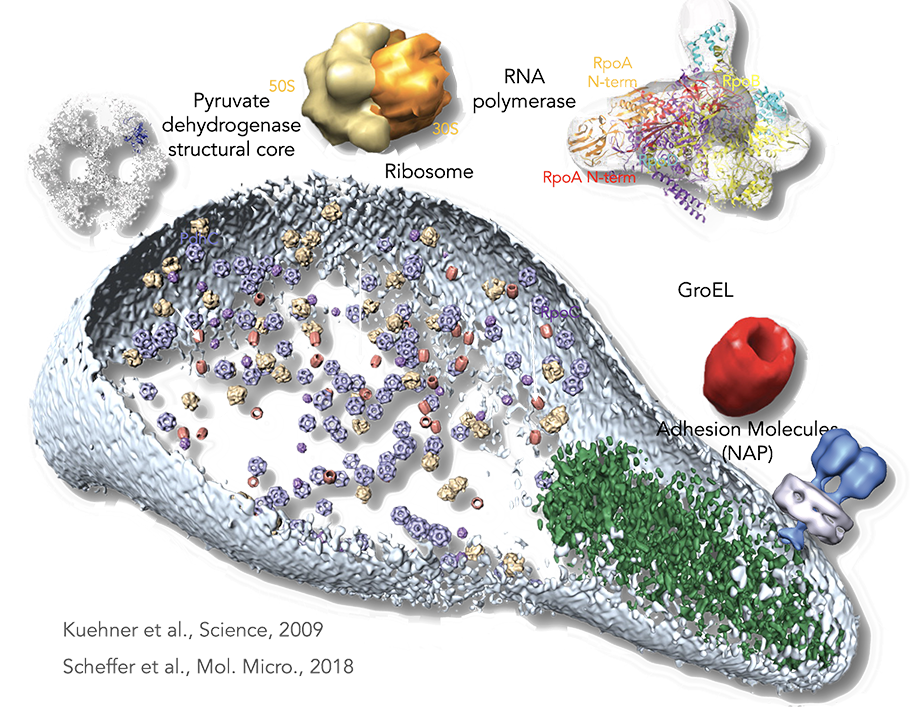
Figure legend: From proteomics to the cell. By a combination of pattern recognition and classification algorithms, the following TAP-identified complexes from M. pneumoniae, matching to existing electron microscopy and x-ray and tomogram structures were placed in a whole-cell tomogram. The structural core of pyruvate dehydrogenase in blue (~23 nm), the ribosome in yellow (~26 nm), RNA polymerase in purple (~17 nm), and GroEL homomultimer in red (~20 nm). Cell dimensions are ~300 nm by 700 nm. The cell membrane is shown in light blue. The rod, a prominent structure filling the space of the tip region, is depicted in green. Its major structural elements are HMW2 (Mpn310) in the core and HMW3 (Mpn452) in the periphery, stabilizing the rod.
In vitro and in situ analysis of the RNA-Polymerase and (Poly)-Ribosomes in bacteria
The direct study of epigenetic events, such as transcription, modification or binding, requires imaging of individual genes at molecular resolution. Electron microscopy (EM) can show local detail of the genome. However, direct EM visualization and analysis of specific individual genes is currently not feasible for two reasons: genes cannot be unambiguously localized in the crowded, landmark-free environment of the nucleus; and the amount of complexes found in the electron microscope is too small for quantitative studies. Our methodologies allows for the insertion and direct visualization of gene clusters for a range of analyses, such as changes in gene activity upon alteration of cellular or external factors.
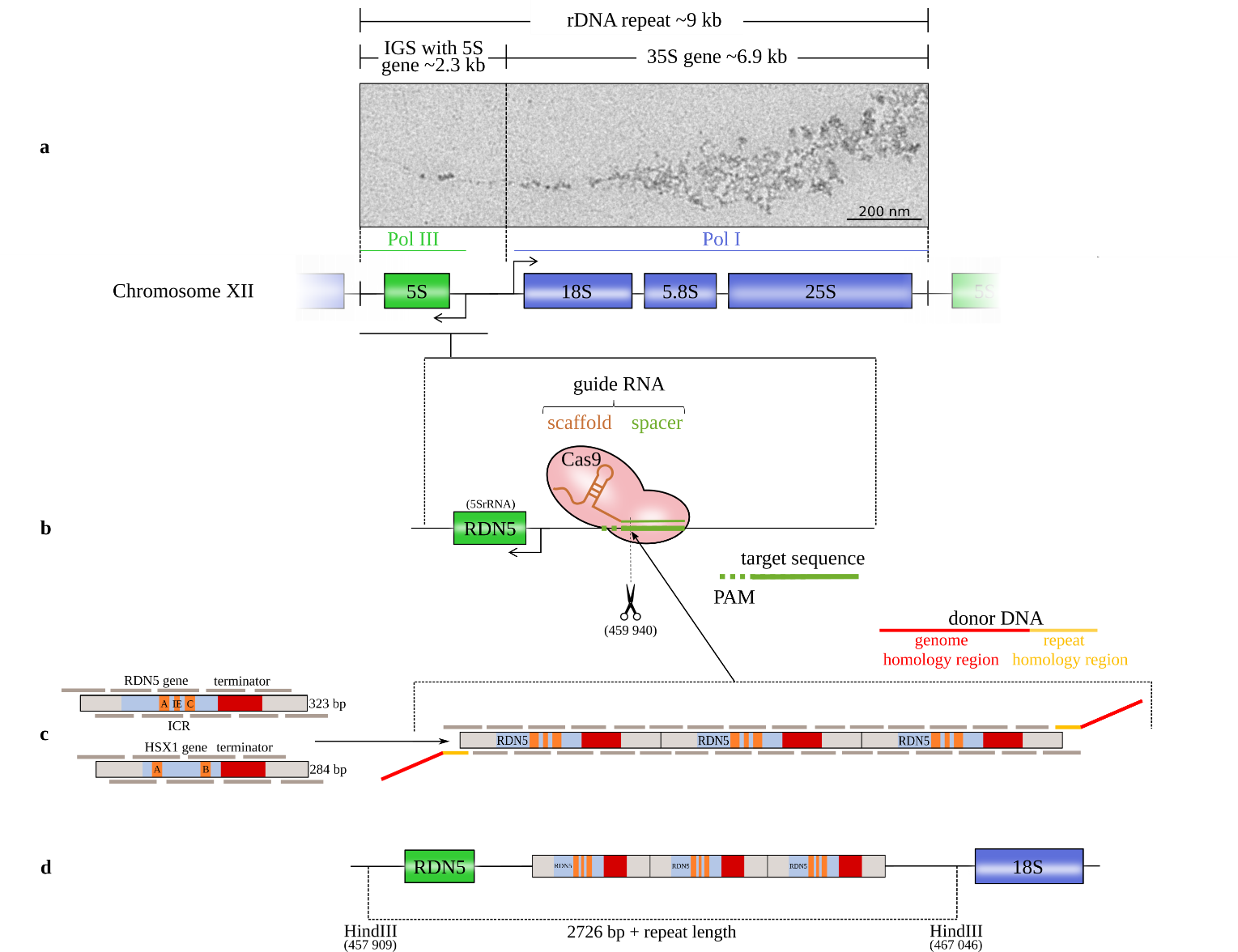
Figure legend: Schematic of the method for introducing gene copies. (a) The rRNA repeats on S. cerevisiae chromosome XII with the 18S, 5.8S, and 25S rRNAs (transcribed by the RNA polymerase I (Pol I)) and the 5S rRNA (transcribed by Pol III in the reverse direction from the complementary strand). The 5S rRNA gene is located in the intergenic spacer (IGS) between successive repeats (Saccharomyces Genome Database standard name: RDN5-1). EM image: Miller tree in chromatin spreads of S. cerevisiae strain IMX672 at 12,000x magnification from positively stained sample. (b) The CRISPR/Cas9 system induces a double-strand break at the chosen target site with a protospacer adjacent motif (PAM) downstream of the RDN5 gene (5S rRNA) within the rRNA repeat. (c) Both copy units cover the gene sequence, the terminator and a 50 bp spacer on both sides. RDN5 is a type 1 promotor gene with an internal control region (ICR) that contains box A and box C elements and an intermediate element (IE). The internal promotor requires the transcription factors IIIC, IIIB and IIIA (TFIIIC, TFIIIB and TFIIIA). HSX1 is a type 2 promotor gene with box A and box B elements that requires only TFIIIC and TFIIIB [8]. The copy units are divided into overlapping oligomers. The in vivo repeat assembly is targeted to the Cas9 cleavage site by donor DNA adapters. (d) After Cas9 cleavage, the copy cluster is inserted at the DSB site.
Deciphering the architecture of cell-cell junctions
Organs are composed of multiple cell types, which require proper intercellular communication for development and homeostasis. The behavior of the cells is strongly influenced by cell-cell junctions, which constitute structural bridges between cells that have critical roles in tissue stability, homeostasis and signaling. The structures of the individual proteins that build up these junctions are largely known, although the molecular arrangement responsible for their many functions remains elusive. The blood-brain barrier (BBB) is the most prominent entity that relies on selective cell-cell junctions acting as a barrier to control the transport of substances across the central nervous system and maintaining brain homeostasis. BBB function is built on a specific arrangement of multiple cell types: endothelial cells form the vascular walls and are surrounded by mural cells (smooth muscle cells and pericytes), both of which are ensheathed in the basal lamina (Fig. 2); and projections of astrocytes, called astrocytic endfeet, encircle the brain vasculature and mediate communication between the vascular and nervous systems. One level, the paracellular route, relies on the proper assembly of endothelial cell junctions (tight junctions and adherens junctions) between adjacent endothelial cells to impair the passage of molecules through the intercellular space (Stamatovic et al., 2016). Defects in cell-cell junction proteins lead to BBB impairment and leakage, which allows the paracellular passage of substances and blood cells – an outcome or an aggravating factor in neuropathologies (Sweeney et al., 2019). Conversely, the possibility of pharmacologically manipulating the opening of the BBB by temporally loosening the endothelial junction assembly could allow therapeutic access to the central nervous system.
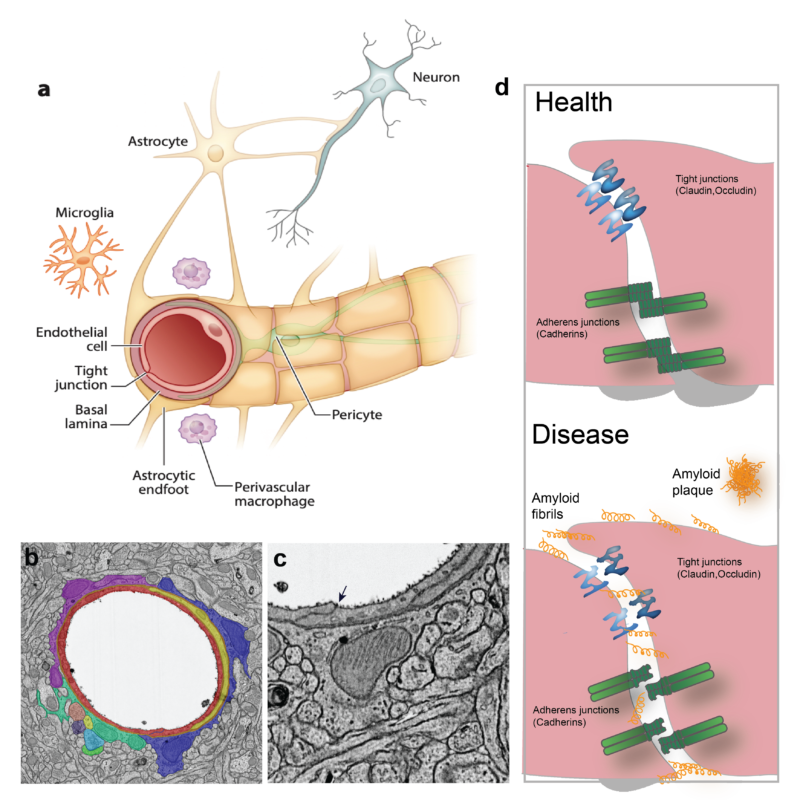
Figure legend: Cellular arrangement in the brain vasculature. a) The brain vasculature has the unique characteristic of being non-permeable to blood-borne substances. This is accomplished by a tight connection between the cells that form the neurovascular unit: endothelial cells, pericytes and astrocytes. Endothelial cells and pericytes are surrounded by an extracellular matrix called the basal lamina. b) Transmission electron microscopy picture of a mouse brain section showing a blood vessel that has been artificially colored to better depict the cellular arrangement in the brain vasculature (red: endothelial cells; yellow: pericyte; purple, blue and green: astrocytic endfeet). Arrow indicates a tight junction between adjacent endothelial cells. c) Magnification of b) showing the tight junction as an electron-dense line sealing the interface between endothelial cells. d) Detailed cartoon of square in a) showing the protein junction arrangement in health and disease conditions. Tight junctions and adherens junctions impair the paracellular passage of molecules into the brain parenchyma and ensure BBB integrity. Although the arrangement of endothelial junctions is known to be altered in pathologies (such as Alzheimer’s disease), the ultrastructural disorganization of junctions in disease remains largely unknown. Figure adapted from (Segarra et al., 2019).
